Researchers using a new genetic screening technique have found seven new tumor-suppressor genes in head and neck cancer (HNC) whose role in cancer was previously unknown, according to a new study in Science (January 17, 2014, Vol. 343:6168, pp. 309-313).
Scientists at Rockefeller University in New York created the new technique, which can eliminate random bystander genes and identify the ones that are critical for cancer.
The technique, which the researchers recently applied to a screen for skin tumor genes, is particularly useful because it is faster and easier than the traditional method for determining gene function -- breeding genetically modified animals to study the impact of missing genes.
"Using knockout mice, which are model organisms bred to have a particular gene missing, is not feasible when there are 800 potential head and neck cancer genes to sort through," explained Daniel Schramek, a postdoctoral fellow at Rockefeller's Laboratory of Mammalian Cell Biology and Development, which is headed by Elaine Fuchs, PhD. "It can take about two years per gene. Our method can assess about 300 genes in a single mouse, in as little as five weeks."
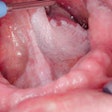
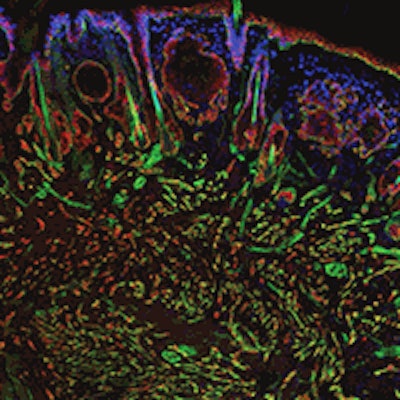
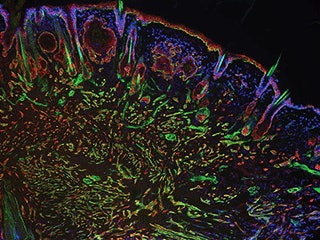 Interfering with cancer. A section of a head and neck tumor -- red and green markers show the proliferation of cancer stem cells -- that formed when one of several newly characterized genes, Myh9, was suppressed. A recently developed genetic screening technique using RNA interference identified Myh9's protein, myosin IIa, as playing an important role in tumor suppression. Image courtesy of Rockefeller University.
Interfering with cancer. A section of a head and neck tumor -- red and green markers show the proliferation of cancer stem cells -- that formed when one of several newly characterized genes, Myh9, was suppressed. A recently developed genetic screening technique using RNA interference identified Myh9's protein, myosin IIa, as playing an important role in tumor suppression. Image courtesy of Rockefeller University.
The researchers used RNA interference, a natural process in which RNA molecules inhibit gene expression. They took short pieces of RNA, which can turn off the function of specific genes, and attached them to highly concentrated viruses. They then used ultrasound to guide the needle without damaging surrounding tissue and injected the viruses into the sacs of mouse embryos.
"The virus is absorbed and integrated into the chromosomes of the single layer of surface cells that cover the tiny embryo," Fuchs said. "As the embryo develops, this layer of cells becomes the skin, mammary glands, and oral tissue, enabling us to efficiently, selectively and quickly eliminate the expression of any desired gene in these tissues. The noninvasive method avoids triggering a wound or inflammatory response that is typically associated with conventional methods to knockdown a gene in cultured cells and then engraft the cells onto a mouse."
As the mice grew, the researchers determined which genes were promoting tumor growth when turned off, and they were surprised at what they found.
“This method of RNA interference is highly useful in the rapid discovery, validation, and characterization of tumor suppressor genes that might otherwise be missed in a genetic screen.”
Rockefeller University
"Among the seven novel tumor suppressor genes we found, our strongest hit was Myh9, which codes for the protein myosin IIa, a motor protein with well-known function in cell structure and cell migration," Schramek said. "Through further functional studies we found that myosin IIa is also required for activation of the main guardian of the genome -- a tumor-suppressor protein called p53."
When the myosin IIa gene was mutated, the researchers found that p53 was not able to build up in the cell nucleus, and chaos ensued: Genes responsible for repairing damaged cells and killing off tumor cells were not activated, and invasive carcinomas spread within three months.
The researchers devised a way to reactivate p53 in these cells and showed in vitro that tumor suppression was restored.
"Head and neck cancers are the sixth most deadly type of cancer worldwide. Interestingly, Myh9 is also mutated in human head and neck cancers, and low expression of myosin IIa correlates with poor prognosis for the patient," Fuchs said.
The researchers will now examine the effect in future clinical trials and will look at the function of the other six genes their study identified.
"We've demonstrated that this method of RNA interference is highly useful in the rapid discovery, validation, and characterization of tumor suppressor genes that might otherwise be missed in a genetic screen," Schramek said. "It can be applied to many kinds of cancers, such as breast and lung."